The Cooper Trewin Foundation is proud to support the Robert’s Program on Sudden Unexpected Death in Pediatrics at Boston Children’s Hospital including their innovative Zebrafish Project, which investigates genetic factors contributing to sudden unexplained deaths in children (SUDC).
We are pleased to share an update from Dr. Richard D. Goldstein, MD, Program Director of Robert’s Program, detailing the progress of the project and the ongoing research into the gene DEPDC5, which may play a critical role in understanding sudden death in pediatric populations.
…………………………………………………………………………………
Letter from Dr. Richard D. Goldstein, MD
25 August 2024
Re: Update on the Robert’s Program Zebrafish Project supported by the Cooper Trewin Memorial SUDC Research Fund.
Dear Rob,
It was a pleasure to meet with you at Boston Children’s a few months ago and show you some of the work being done by members of our team working on our project, entitled Modelling Genetic Findings in Sudden Unexpected Death in Pediatrics (SUDP) in Zebrafish, or the Cooper Trewin Zebrafish Project.
Our team continues to make progress both in terms of generating genetic zebrafish models that are directly relevant to SUDP and characterizing a primary phenotypic feature in the fish that we sought to describe, which is decreased survival. It is not always the case that a genetic animal model will recapitulate what is seen in humans. In this case, simply put, our initial larval zebrafish models of the gene DEPDC5 die prematurely, which establishes important ties between the gene and the tragic, untimely premature death of children dying from SUDP. We will review some of the experimental data below. We have achieved an excellent first step in modelling, and to understand the mechanisms underlying this premature death, we are bringing in more sophisticated ways of generating ‘mosaic’ models that will survive longer because not all of the cells in the animals harbor the ‘mutant’ gene. Further studies of mosaic DEPDC5 models form the basis of our next phase of research, as outlined in the accompanying proposal.
To review our background and rationale for conducting the current project, our human genetics (DNA sequencing) studies in 352 of children whose deaths were due to SIDS and SUDC implicated a number of genes related to neurological, cardiac, and other disorders (published in Koh et al., Genetics in Medicine, 2022). The Cooper Trewin Zebrafish Project (phase 1) focused on genes involved in actual SUDC cases that are expressed in the brain and related to neurological conditions like epilepsy. We altered these genes by inserting CRISPR gene editing constructs into zebrafish one-cell embryos and characterizing the resulting mutant (“crispant”) larvae. Our phenotyping methods include evaluation of survival, quantification of brain size, detection of seizure-like swimming behavior, electrophysiology of the zebrafish brain, and assays of behavioral correlates of anxiety as well as learning.
We spent the initial months of the project generating germline models targeting the genes SCN1A and DEPDC5, long known and important causes of epilepsy that are associated with Sudden Unexpected Death in the Epilepsy (SUDEP) and found recently in SIDS and SUDC through research from our group and others. We started with these neurologically focused experiments and had such robust success in studying DEPDC5 that we have maintained focus on this gene and the associated pathway, which is the mammalian target of rapamycin, or mTOR, pathway. We will focus on the characterization of this gene’s models in this update. We had earlier presented preliminary data on models of SCN1A, another gene that we and others have reported to be involved in SUDP as well as Sudden Unexplained Death in Epilepsy. The phenotypes of these models, though previously published to display features of epilepsy, have not been as striking from a SUDP standpoint. (We are investigating why we and other labs are not seeing seizures in this model in a separate study in the Poduri Lab.) We had also originally planned to evaluate potential cardiac effects of disrupting these genes, which we will continue to prioritize as we generate these models.
DEPDC5 studies: As noted in our interim report in 2022, we demonstrated amino acid sequence homology between human DEPDC5 vs. zebrafish Depdc5. The dotted blue line with arrowheads denotes a highly conserved region, in which we had observed a variant in one of the probands in our human genetics study (Koh et al., Genetics in Medicine, 2021). Our zebrafish guide RNA sequence and center of the target site is denoted by the red arrow.
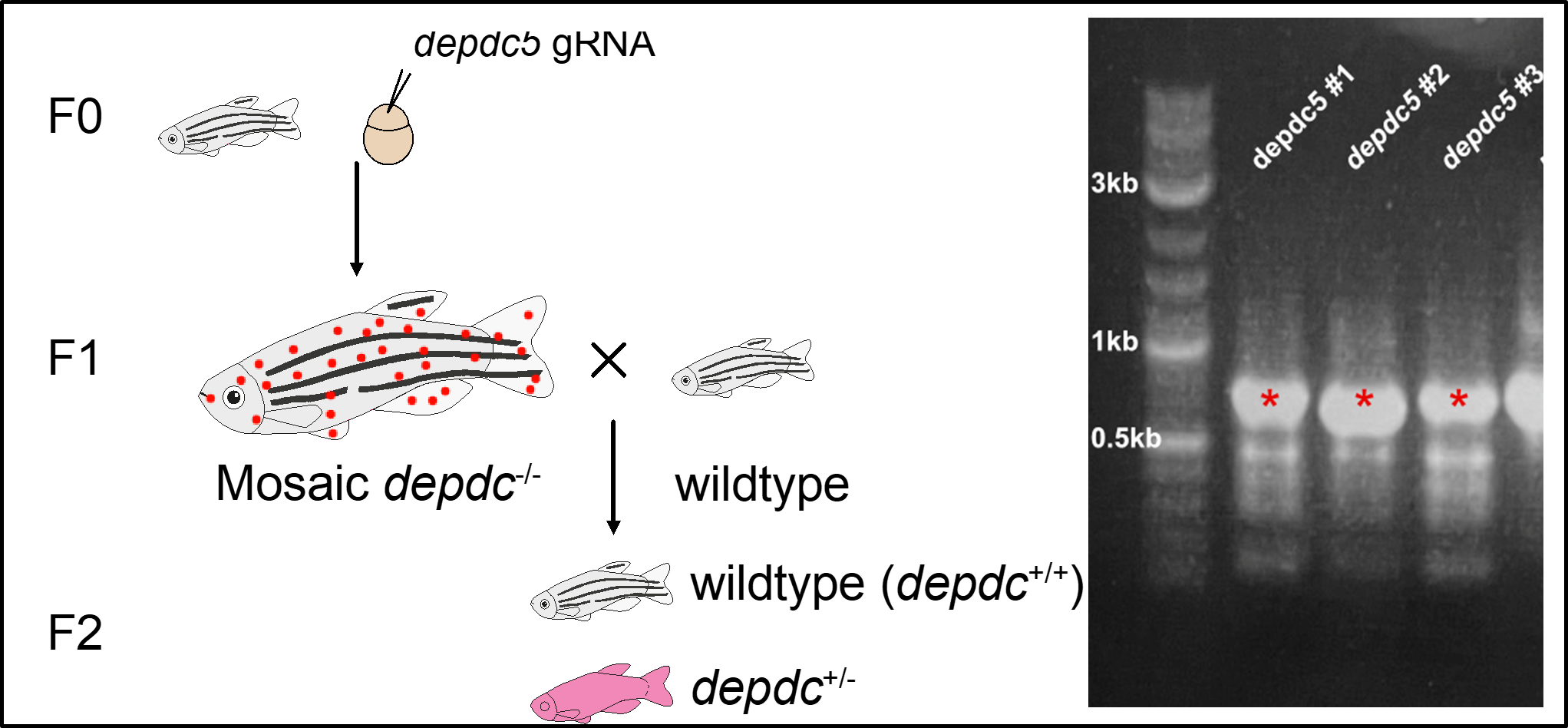
After injecting the above CRISPR guide (sgRNA) directed at zebrafish Depdc5 into 1-cell zebrafish embyros (see figure below), we assessed survival of the mutant larvae (crispants, F0 generation). In this initial round of experiments, survival appeared intact. These F0 crispants are “mosaic” in that some cells have variants and some do not due to less than 100% efficiency of the CRISPR/Cas9 system. We bred these fish with each other to generate the F1 generation, some of which would have carried the variant form, assuming the gametes of the parents had acquired the variant. The F1 mosaic larvae were assessed by Western blot and found to still express some Depdc5 protein. Assuming that some zebrafish would have the wild type (wt) and some the crispant form of Depdc5, we have further crossed them to wt fish to generate germline mutants in the F2 generation. Crosses between mutant fish generate larvae for survival assays and neurological phenotyping, performed blind to genotype and then followed by genotyping. The genotyping strategy and Western blot are shown below.
DEPDC5 survival studies
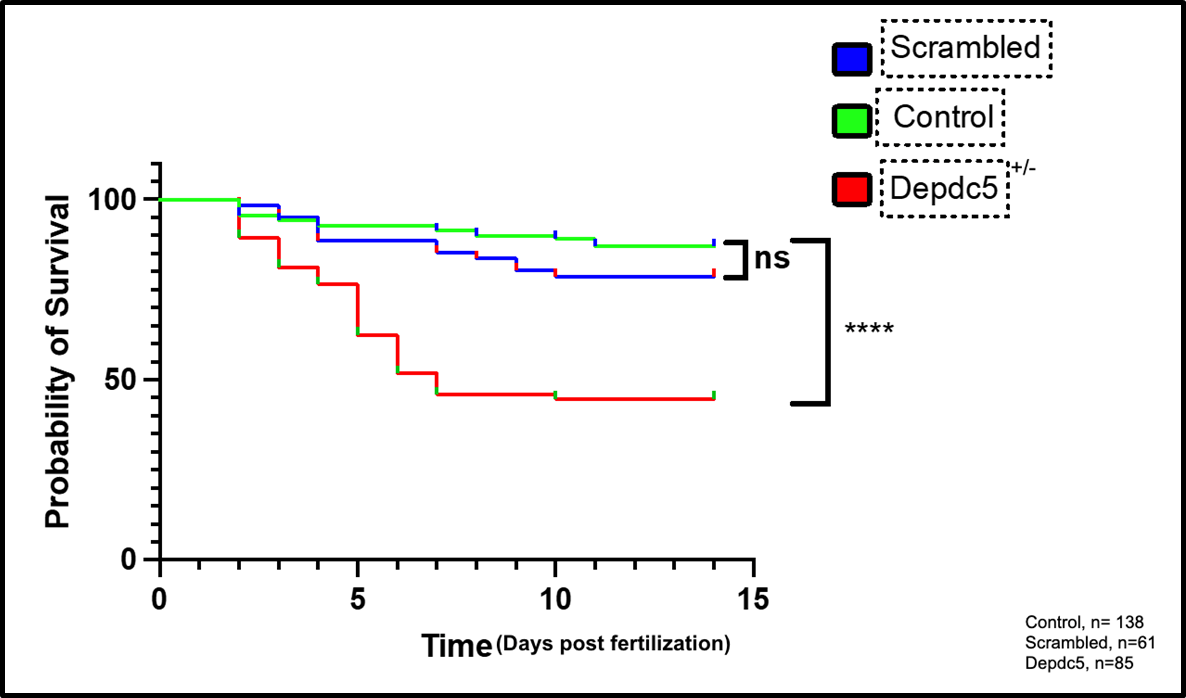
Using the F2 and subsequent generations of depdc5 crispants, we generate larvae through het x het crosses, which gives us an abundance of heterozygous depdc5 (+/-) larvae and wt controls from the same clutches. We also generate scrambled, or sham, crispants, targeting genes that are non-essential (“housekeeping” genes), as controls who have undergone the processes of injection, transfer of guide RNAs, exposure to Cas9, etc. Strikingly, when we compare survival of larvae that are depdc5 (+/-) mutant vs. wt same-clutch controls vs. scrambled controls, we see an early loss of survival phenotype in the depdc5 (+/-) mutants alone, as demonstrated in the figure to the right.
These are important steps in understanding the vulnerability of some infants and children to SUDP. We have also made important strides forward in the development of an experimental model that may help us better understand the mechanisms of early death. Ultimately, we hope that methods to prevent death and understand risk will come from this greater understanding of these mechanisms, though we know we have a long way to go.
Impact on the next generation of scientists: Other positive outcomes of this study have included engaging post-doctoral fellow Dr. Sneham Tiwari in the research, who joined the Poduri Lab shortly before our former post-doctoral fellow starting his residency in Child Neurology at Baylor College of Medicine. Dr. Tiwari was immediately interested in investigating the function of DEPDC5 and other genes related to SUDP and/or seizures, and she obtained a fellowship grant to support her time working the rest of the team on generating and studying the animal models. Likewise, the entire lab team has recognized the importance of considering death as a phenotype in all of the genetic epilepsy modeling that we do. Dr. Tiwari presented her preliminary results from this project at the prestigious Gordon Research Conference on Epilepsy and Mechanisms of Neuronal Synchronization this month.
We continue to be grateful for the generous support of the Cooper Trewin Memorial SUDC Research Fund. We hope you are as excited as we are that this work is moving us toward a deeper understanding of genetic factors that can predispose children to SUDP. We are most excited to take on the next phase of research that will build on the foundation described above, and we have summarized our next set of experiments in the accompanying proposal. We sincerely hope that today’s understanding will lead to tomorrow’s prevention of SIDS and SUDC. Thank you so very much.
Sincerely,
Richard D. Goldstein, MD
Program Director, Robert’s Program
Annapurna Poduri, MD, MPH
Principal Investigator, Poduri Lab and Epilepsy Genetics Program
Neurologist, Robert’s Program


Recent Comments